Generate tab
Shiver uses multi-dimensional event workspaces as an intermediate data storage. These are
neutron events labeled by the reciprocal space coordinates. Once all the parameters are set and
valid, one can click on the Generate button, to create and save an MD event workspace. Once
the process is done, the workspace will appear in the list in the Main window. One can
also Save configuration as a python file.
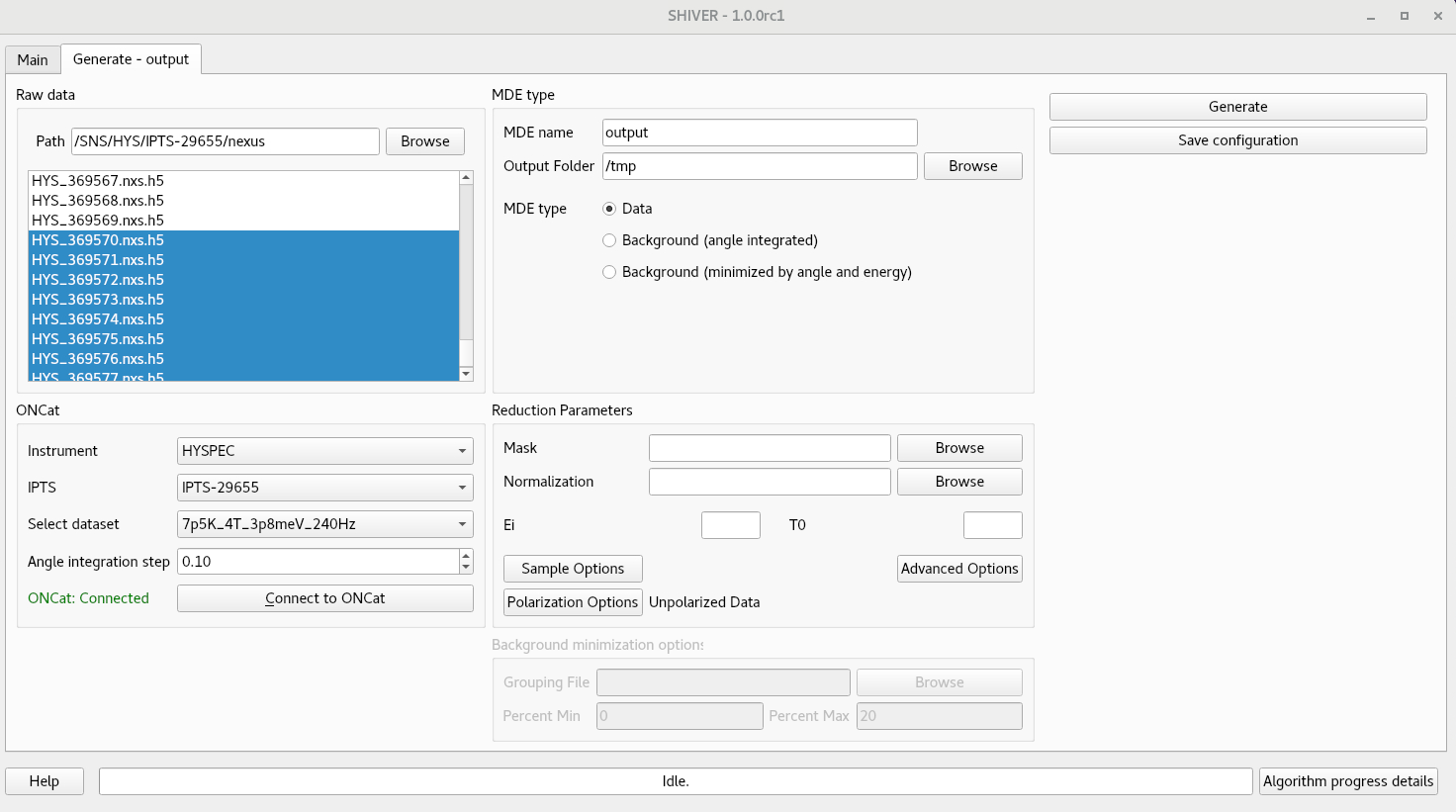
Raw data files
There are two ways to choose the raw data files. The standard way is to navigate to the folder, then
click (or shift+click, or ctrl+click, as appropriate) to select. The second option, if the appropriate
metadata was included during acquisition, is to use the ONCat database.
- Make sure you are connected to the database. Use UCAMS/XCAMS and password
- Select instrument
- The available experiment (IPTS) numbers is repopulated. Select one
- The name of available datasets is prepopulated.
This is written as sequence name in the raw file. For some instruments
datasets are labeled by the comments instead. If that is the case, one needs to modify
the
~/.shiver/configuration.inifile, and setuse_notestoTrue. If dataset names are not available, or one changes manually the selection, theSelect datasetfield will change tocustom - Select an angle integration step. Runs within the same step will be grouped together. This can reduce memory size, if several measurements were done at the same angle.
- The omega angle is used as a default value for grouping the datafiles.
The value can be updated from the
Advanced Optionsdialog ->Goniometerfield. When the changes are saved, they are immediately updated in the oncat view. - The use_notes configuration variable is read and used on every field dropdown change.
The variable can be configurered from the
Configuration Settingsdialog.
Data type and location
The user must enter a name for the workspace to be created, and a location to save the file. The file
name will be the same as the workspace name, plus the .nxs extension.
There are three types of processing:
datais the standard way, when each event is labeled by the momentum transfer in the goniometer frame. Workspaces of this type will show up in the main window with labelQS, and can be selected as data or background. An example of using this angle dependent background can be the case when one wants to subtract two different temperatures, measured at the same angles.Background (angle integrated)adds together all the selected runs and ignores the goniometer value. This process can be used if one measures an empty can type of background. The output workspace will show up in the main window with labelQL, and can only be used as a backgroundBackground (minimized by angle and energy)is a new way of calculating a powder-like background directly from the data. Nearby detectors are grouped together, and events are then binned in energy transfer. For each detector group and energy bin, one calculates then sorts the intensity for each sample rotation. The user then selects runs with the intensities in a desired percentage range. The parameters for this option,Grouping file,Percent Min, andPercent Max, can be found in theBackground minimization optionsarea. Ask the local contact for a good grouping file.
Reduction parameters
Users can specify additional reduction parameters. A Mask file is a processed Mantid NeXus file.
The Normalization file can also contain a mask, but it also contains the efficiency of the
detectors, measured using some incoherent scatterer (usually Vanadium or TiZr).
The incident energy Ei and the time offset at the moderator T0 are automatically calculated
from the monitors or from metadata in the file. However, if these are incorrect, one can
just override them.
The Sample options is required to set up lattice parameters and orientation.
The Polarization Options is used for HYSPEC measurements to set polarization state, flipping ratio, and, optionally, to overide the status of the supermirror analyzer.